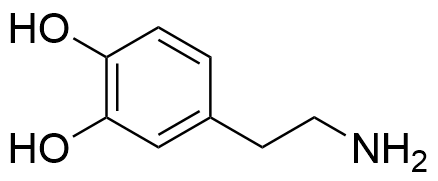
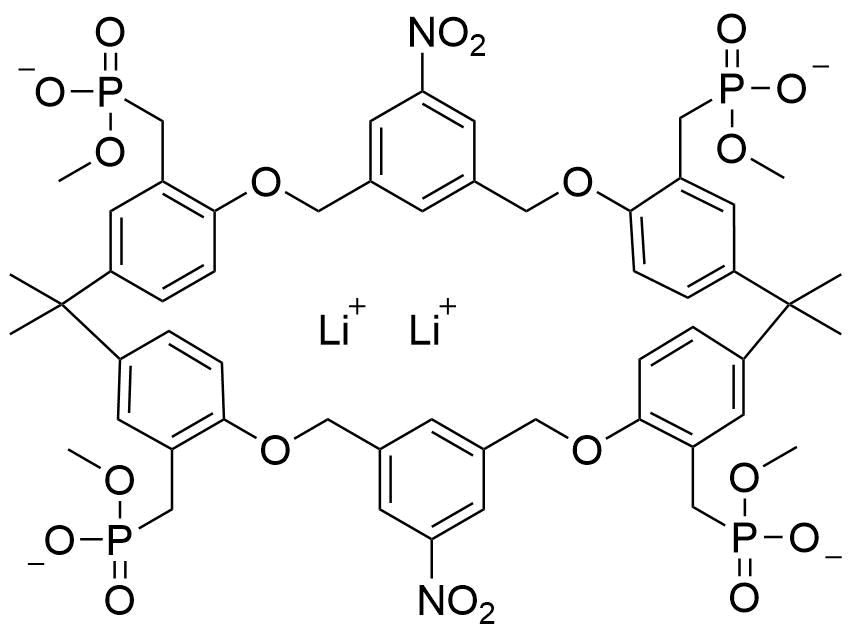
Binding Properties
| 𝜈 | Molecule 1 : 1 Host | ||
| Ka = | 246.0 | ± 93.5 | M-1 |
| Kd = | |||
| logKa = | |||
| T | 25.0 °C | ||
| Energy | kJ mol-1 | kcal mol-1 | |||
|---|---|---|---|---|---|
| ΔG | = | -13.65 | ± 0.99 | -3.26 | ± 0.24 |
These are the specifications of the determination of the experimental results.
| Detection Method: | Direct | ||
| Assay Type: | Direct Binding Assay | ||
| Technique: | Nuclear Magnetic Resonance | ||
| = | 0.2 ppm | ||
Detailed information about the solvation.
| Solvent System | Complex Mixture | |
| Solvents | Deuterium Oxide | 50.0 % |
| deuterated meth... | 50.0 % | |
Please find here information about the dataset this interaction is part of.
| Citation: |
T. Schrader, M. Herm, O. Molt, SupraBank 2024, Towards Synthetic Adrenaline Receptors—Shape-Selective Adrenaline Recognition in Water (dataset). https://doi.org/10.34804/supra.20210928204 |
| Link: | https://doi.org/10.34804/supra.20210928204 |
| Export: | BibTex | RIS | EndNote |
Please find here information about the scholarly article describing the results derived from that data.
| Citation: |
M. Herm, O. Molt, T. Schrader, Angew. Chem. Int. Ed. 2001, 40, 3148–3151. |
| Link: | https://doi.org/10.1002/1521-3773(20010903)40:17%3C3148::AID-ANIE3148%3E3.0.CO;2-S |
| Export: | BibTex | RIS | EndNote |
Binding Isotherm Simulations
The plot depicts the binding isotherm simulation of a 1:1 interaction of Dopamine (0.08130081300813008 M) and Tetramethyl((2,2,10,10-tetramethyl-65,145-dinitro-4,8,12,16-tetraoxa-1,3,9,11(1,4),6,14(1,3)-hexabenzenacyclohexadecaphane-13,33,92,113-tetrayl)tetrakis(methylene))tetrakis(phosphonate) (0 — 0.16260162601626016 M).
Please sign in: customize the simulation by signing in to the SupraBank.