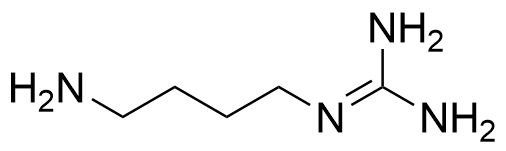
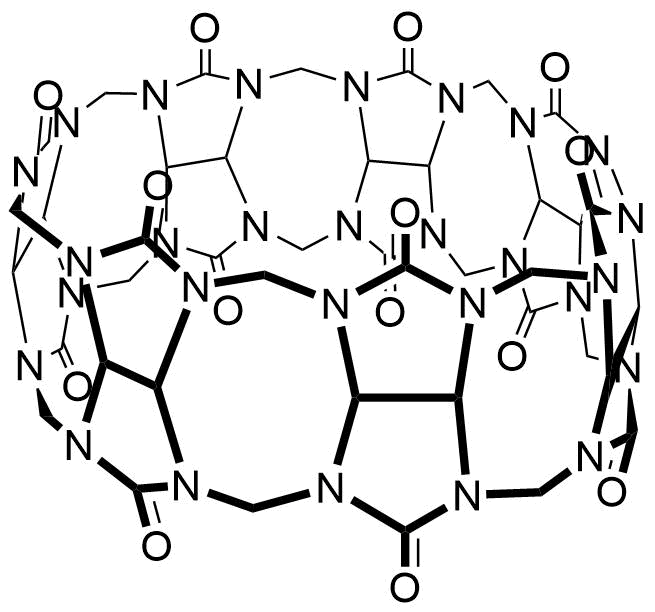
Binding Properties
| 𝜈 | Molecule 1 : 1 Host | ||
| Ka = | 1.10⋅106 | ± 1.10⋅105 | M-1 |
| Kd = | |||
| logKa = | |||
| T | 25.0 °C | ||
| Energy | kJ mol-1 | kcal mol-1 | |||
|---|---|---|---|---|---|
| ΔG | = | -34.48 | ± 0.25 | -8.24 | ± 0.06 |
These are the specifications of the determination of the experimental results.
| Detection Method: | Competitive | |||
| Assay Type: | Competitive Binding Assay | |||
| Technique: | Fluorescence | |||
| 𝛌ex | = | 336.0 nm | ||
| 𝛌em | = | 380.0 nm | ||
Detailed information about the solvation.
| Solvent System | Buffer System | 10 mM ammonium acetate pH-6.0 |
| Solvents | water | 100.0 % |
| Additives | Ammonium acetate | 10.0 mM |
| Source of Concentration | ||
| Total concentration | 10.0 mM | |
| pH | 6.0 |
Please find here information about the dataset this interaction is part of.
| Citation: |
W. M. Nau, V. D. Uzunova, A. Hennig, D. M. Bailey, SupraBank 2024, Supramolecular Tandem Enzyme Assays for Multiparameter Sensor Arrays and Enantiomeric Excess Determination of Amino Acids (dataset). https://doi.org/10.34804/supra.20210928260 |
| Link: | https://doi.org/10.34804/supra.20210928260 |
| Export: | BibTex | RIS | EndNote |
Please find here information about the scholarly article describing the results derived from that data.
| Citation: |
D. M. Bailey, A. Hennig, V. D. Uzunova, W. M. Nau, Chem. Eur. J. 2008, 14, 6069–6077. |
| Link: | https://doi.org/10.1002/chem.200800463 |
| Export: | BibTex | RIS | EndNote |
Binding Isotherm Simulations
The plot depicts the binding isotherm simulation of a 1:1 interaction of Agmatine (1.8181818181818182e-05 M) and CB7 (0 — 3.6363636363636364e-05 M).
Please sign in: customize the simulation by signing in to the SupraBank.