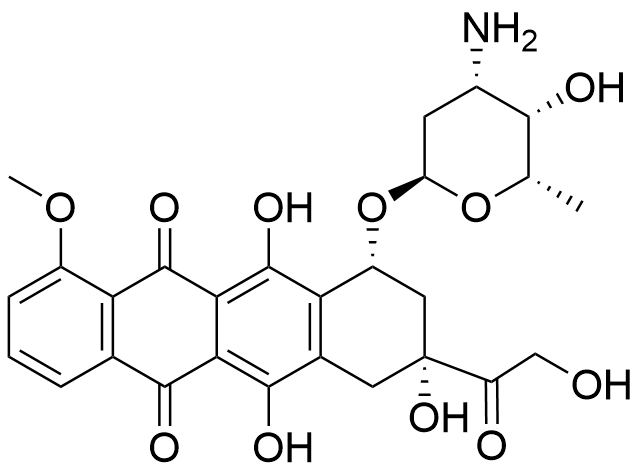
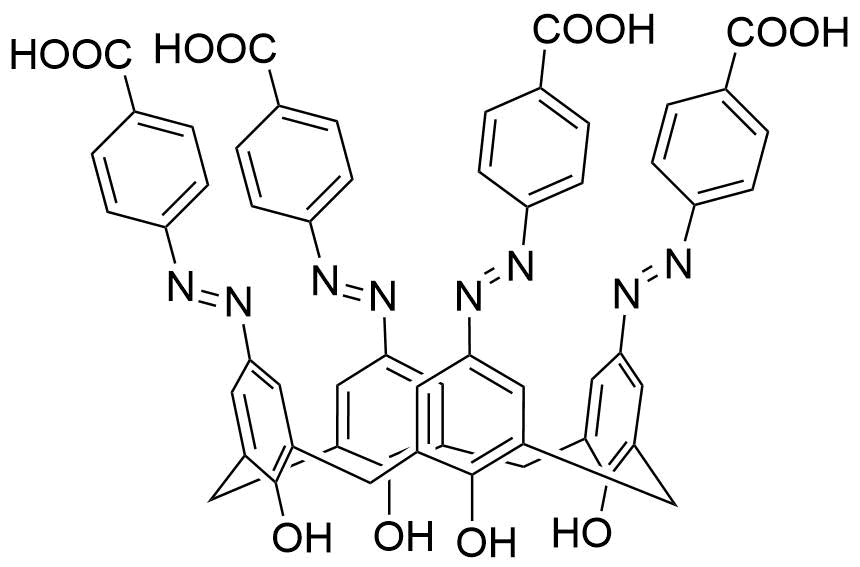
Binding Properties
| 𝜈 | Molecule 1 : 1 Host | ||
| Ka = | 1.60⋅108 | ± 5.00⋅107 | M-1 |
| Kd = | |||
| logKa = | |||
| T | 25.0 °C | ||
| Energy | kJ mol-1 | kcal mol-1 | |||
|---|---|---|---|---|---|
| ΔG | = | -46.83 | ± 0.8 | -11.19 | ± 0.19 |
These are the specifications of the determination of the experimental results.
| Detection Method: | Direct | |||
| Assay Type: | Direct Binding Assay | |||
| Technique: | Fluorescence | |||
| 𝛌ex | = | 540.0 nm | ||
| 𝛌em | = | 594.0 nm | ||
Detailed information about the solvation.
| Solvent System | Buffer System | 10 mM PBS pH-7.4 |
| Solvents | water | |
| Additives | sodium chloride | 136.99 mM |
| Disodium hydrog... | 6.13 mM | |
| Sodium dihydrog... | 5.03 mM | |
| potassium chloride | 2.7 mM | |
| Source of Concentration | estimated | |
| Total concentration | 10.0 mM | |
| pH | 7.4 |
Please find here information about the dataset this interaction is part of.
| Citation: |
D. Guo, T. Zhang, Z. Zhang, Y. Yue, X. Hu, F. Huang, L. Shi, Y. Liu, SupraBank 2024, A General Hypoxia‐Responsive Molecular Container for Tumor‐Targeted Therapy (dataset). https://doi.org/10.34804/supra.2021092885 |
| Link: | https://doi.org/10.34804/supra.2021092885 |
| Export: | BibTex | RIS | EndNote |
Please find here information about the scholarly article describing the results derived from that data.
| Citation: |
T. X. Zhang, Z. Z. Zhang, Y. X. Yue, X. Y. Hu, F. Huang, L. Shi, Y. Liu, D. S. Guo, Adv. Mater. 2020, 32, 1908435. |
| Link: | https://doi.org/10.1002/adma.201908435 |
| Export: | BibTex | RIS | EndNote |
Binding Isotherm Simulations
The plot depicts the binding isotherm simulation of a 1:1 interaction of doxorubicin (1.25e-07 M) and aCx4-Bz-COOH (0 — 2.5e-07 M).
Please sign in: customize the simulation by signing in to the SupraBank.