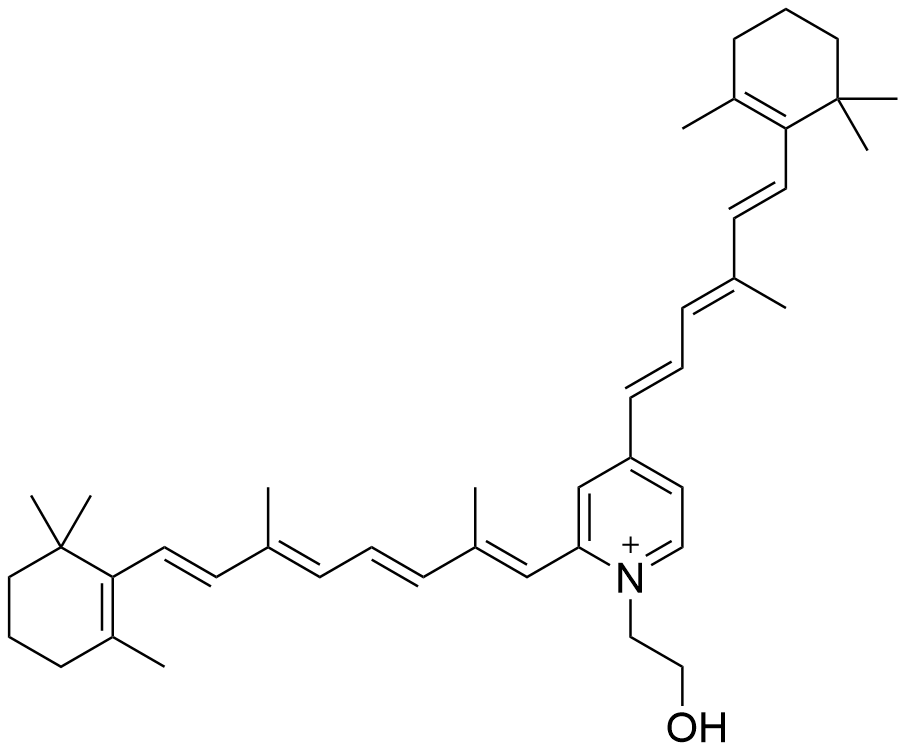
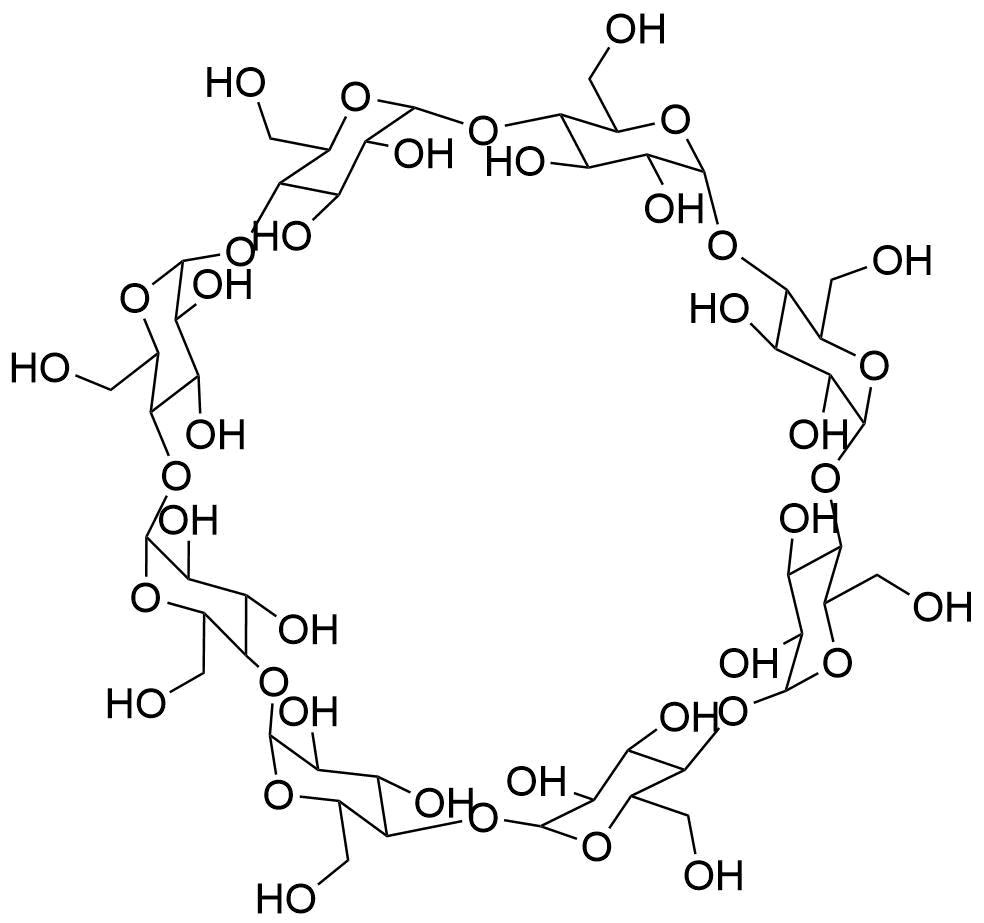
Binding Properties
| 𝜈 | Molecule 1 : 1 Host | ||
| Ka = | 2.74 | M-1 | |
| Kd = | |||
| logKa = | |||
| T | 25.0 °C | ||
| Energy | kJ mol-1 | kcal mol-1 | |||
|---|---|---|---|---|---|
| ΔG | = | -2.5 | -0.6 |
| Detection Method: | Direct | |||
| Assay Type: | Direct Binding Assay | |||
| Technique: | Fluorescence | |||
| 𝛌ex | = | 434.0 nm | ||
| 𝛌em | = | 600.0 nm | ||
| Solvent System | Single Solvent |
| Solvent | water |
| Citation: |
M. M. Nociari, G. L. Lehmann, A. E. Perez Bay, R. A. Radu, Z. Jiang, S. Goicochea, R. Schreiner, J. D. Warren, J. Shan, S. Adam de Beaumais, M. Ménand, M. Sollogoub, F. R. Maxfield, E. Rodriguez-Boulan, SupraBank 2024, Beta cyclodextrins bind, stabilize, and remove lipofuscin bisretinoids from retinal pigment epithelium (dataset). https://doi.org/10.34804/supra.20220628440 |
| Link: | https://doi.org/10.34804/supra.20220628440 |
| Export: | BibTex | RIS | EndNote |
| Citation: |
M. M. Nociari, G. L. Lehmann, A. E. Perez Bay, R. A. Radu, Z. Jiang, S. Goicochea, R. Schreiner, J. D. Warren, J. Shan, S. Adam de Beaumais, et al., Proceedings of the National Academy of Sciences 2014, 111, E1402–E1408. |
| Link: | https://doi.org/10.1073/pnas.1400530111 |
| Export: | BibTex | RIS | EndNote |
Binding Isotherm Simulations
The plot depicts the binding isotherm simulation of a 1:1 interaction of N-retinylidine-N-ethanolamine (7.2992700729927 M) and γ-CD (0 — 14.5985401459854 M).