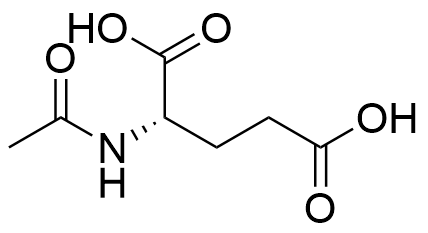
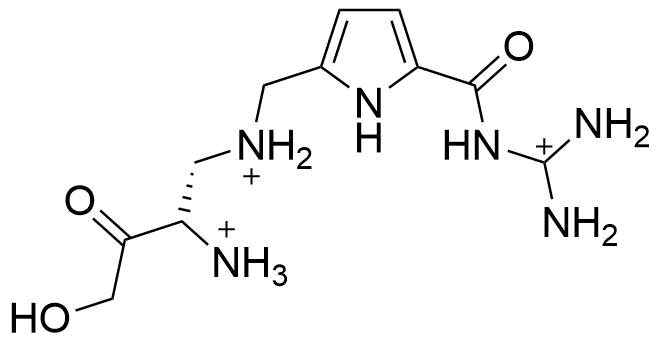
Binding Properties
| 𝜈 | Molecule 2 : 1 Host | ||
| Ka = | 1.52⋅106 | M-2 | |
| Kd = | |||
| logKa = | |||
| T | 25.0 °C | ||
| Energy | kJ mol-1 | kcal mol-1 | |||
|---|---|---|---|---|---|
| ΔG | = | -35.28 | -8.43 |
These are the specifications of the determination of the experimental results.
| Detection Method: | Direct | ||
| Assay Type: | Direct Binding Assay | ||
| Technique: | Nuclear Magnetic Resonance | ||
| Nucleus | H-1 | ||
Detailed information about the solvation.
| Solvent System | Complex Mixture | |
| Solvents | water | 90.0 % |
| dimethyl sulfoxide | 10.0 % | |
Please find here information about the dataset this interaction is part of.
| Citation: |
C. Schmuck, L. Geiger, SupraBank 2024, Efficient Complexation of N-Acetyl Amino Acid Carboxylates in Water by an Artificial Receptor: Unexpected Cooperativity in the Binding of Glutamate but Not Aspartate (dataset). https://doi.org/10.34804/supra.20210928345 |
| Link: | https://doi.org/10.34804/supra.20210928345 |
| Export: | BibTex | RIS | EndNote |
Please find here information about the scholarly article describing the results derived from that data.
| Citation: |
C. Schmuck, L. Geiger, J. Am. Chem. Soc. 2005, 127, 10486–10487. |
| Link: | https://doi.org/10.1021/ja052699k |
| Export: | BibTex | RIS | EndNote | |
Binding Isotherm Simulations
The plot depicts the binding isotherm simulation of a 1:1 interaction of N-Acetyl-L-glutamic acid (1.3175230566534915e-05 M) and (S)-N-diaminomethylcarbamoyl-1H-pyrrol-2-yl-methyl-3-methoxy-3-oxopropane-1,2-diaminium (0 — 2.635046113306983e-05 M).
Please sign in: customize the simulation by signing in to the SupraBank.