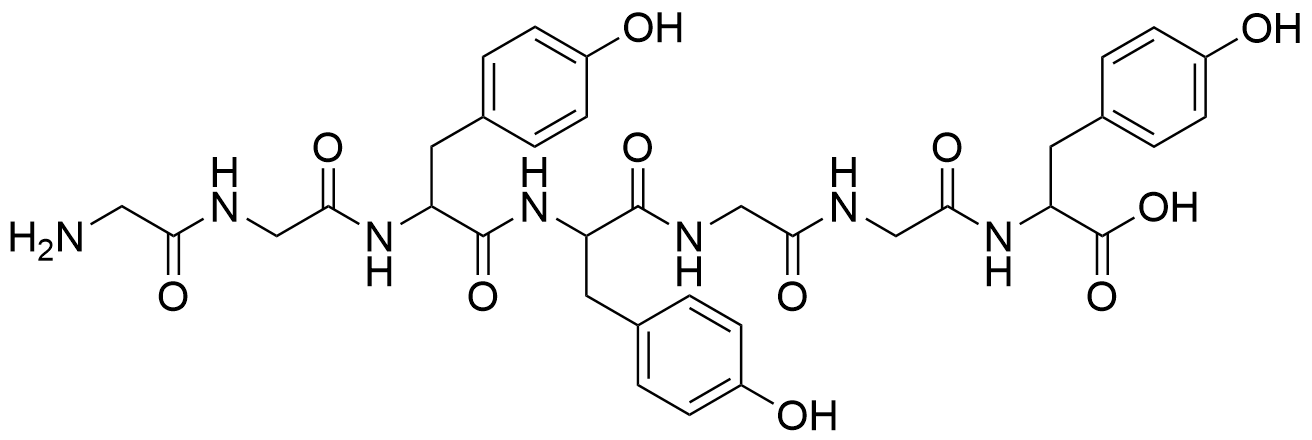
Binding Properties
| 𝜈 | Molecule 1 : 1 Host | ||
| Ka > | 1000.0 | M-1 | |
| Kd > | |||
| logKa > | |||
| T | 27.0 °C | ||
| Energy | kJ mol-1 | kcal mol-1 | |||
|---|---|---|---|---|---|
| ΔG | < | -17.24 | -4.12 |
These are the specifications of the determination of the experimental results.
| Detection Method: | Direct | ||
| Assay Type: | Direct Binding Assay | ||
| Technique: | Isothermal Titration Calorimetry | ||
| Instrument: | Nano ITC | ||
| VCell | = | 1000.0 𝜇L | |
| VSyringe | = | 250.0 𝜇L | |
| Molecule: | syringe | ||
| Partner: | cell | ||
Detailed information about the solvation.
| Solvent System | Buffer System | 10 mM phosphate pH-7.0 |
| Solvents | water | |
| Additives | Sodium dihydrog... | 10.0 mM |
| Sodium hydroxide | ||
| Source of Concentration | ||
| Total concentration | 10.0 mM | |
| pH | 7.0 |
Please find here information about the dataset this interaction is part of.
| Citation: |
P. Suating, C. W. Bielawski, A. Urbach, L. B. Kimberly, M. Ewe, S. L. Chang, J. M. Fontenot, P. R. Sultane, D. A. Decato, O. Berryman, A. Taylor, SupraBank 2024, Cucurbit[8]uril Binds Nonterminal Dipeptide Sites with High Affinity and Induces a Type II β-Turn (dataset). https://doi.org/10.34804/supra.20240228509 |
| Link: | https://doi.org/10.34804/supra.20240228509 |
| Export: | BibTex | RIS | EndNote |
Please find here information about the scholarly article describing the results derived from that data.
| Citation: |
P. Suating, L. B. Kimberly, M. B. Ewe, S. L. Chang, J. M. Fontenot, P. R. Sultane, C. W. Bielawski, D. A. Decato, O. B. Berryman, A. B. Taylor, et al., J. Am. Chem. Soc. 2024, DOI 10.1021/jacs.3c14045. |
| Link: | https://doi.org/10.1021/jacs.3c14045 |
| Export: | BibTex | RIS | EndNote | |
Binding Isotherm Simulations
The plot depicts the binding isotherm simulation of a 1:1 interaction of GGYYGGY Heptapeptide (0.02 M) and CB8 (0 — 0.04 M).
Please sign in: customize the simulation by signing in to the SupraBank.