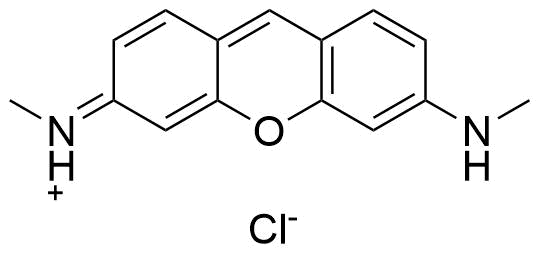
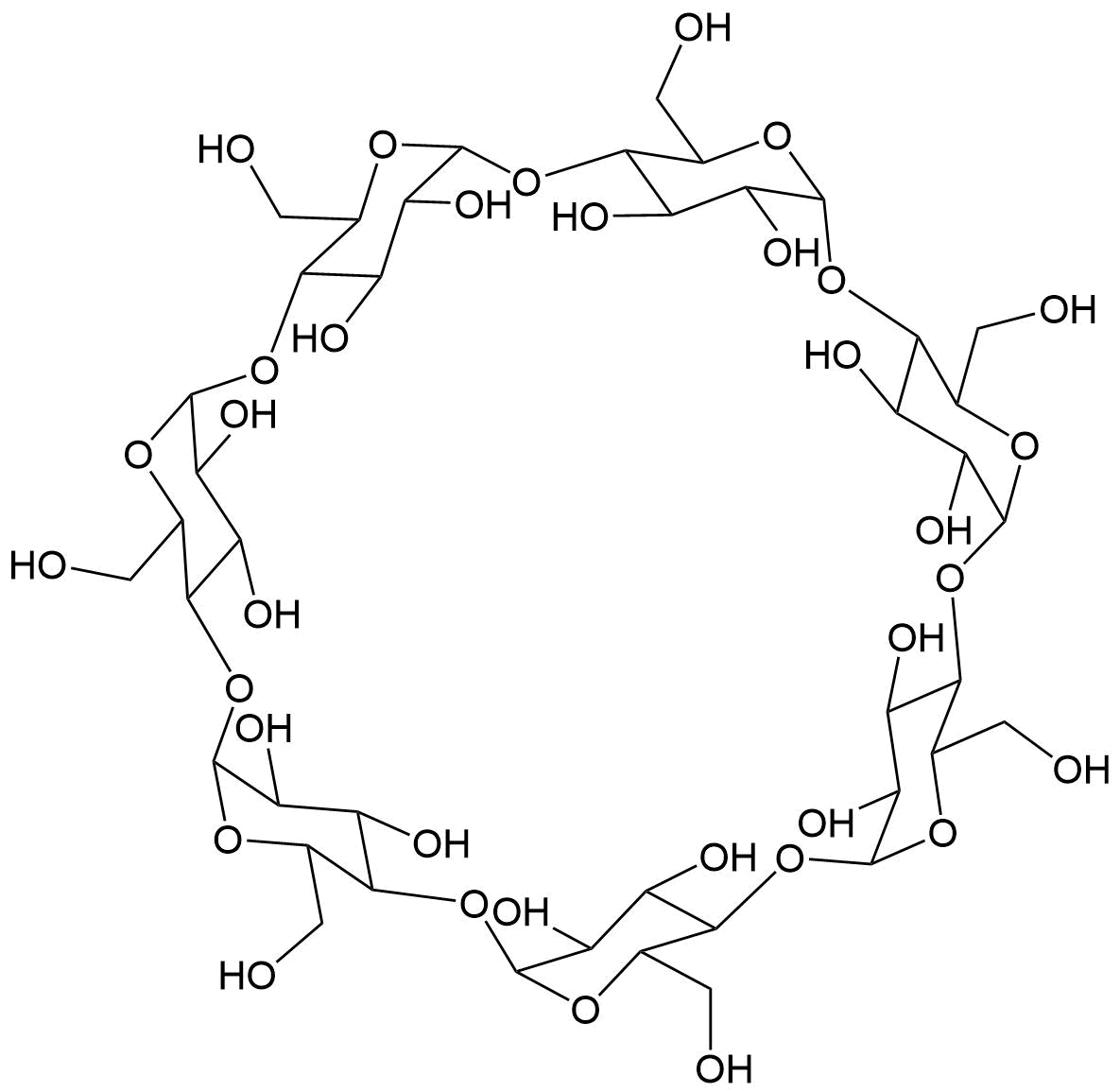
Binding Properties
| 𝜈 | Molecule 1 : 1 Host | ||
| Ka = | 2630.0 | ± 131.5 | M-1 |
| Kd = | |||
| logKa = | |||
| T | 25.0 °C | ||
| Energy | kJ mol-1 | kcal mol-1 | |||
|---|---|---|---|---|---|
| ΔG | = | -19.52 | ± 0.12 | -4.67 | ± 0.03 |
These are the specifications of the determination of the experimental results.
| Detection Method: | Direct | |||
| Assay Type: | Direct Binding Assay | |||
| Technique: | Fluorescence | |||
| 𝛌ex | = | 490.0 nm | ||
| 𝛌em | = | 553.0 nm | ||
| Ibound⁄Ifree | = | 4.0 | ||
Detailed information about the solvation.
| Solvent System | Buffer System | 100 mM phosphate pH-7.2 |
| Solvents | water | |
| Additives | Disodium hydrog... | 66.6 mM |
| Sodium dihydrog... | 33.4 mM | |
| Source of Concentration | estimated | |
| Total concentration | 100.0 mM | |
| pH | 7.2 |
Please find here information about the dataset this interaction is part of.
| Citation: |
C. Hu, SupraBank 2026, Inclusion complexation of β-cyclodextrin and 6-O-α-maltosyl- and 2-O-(2-hydroxypropyl)-β-cyclodextrins with some fluorescent dyes (dataset). https://doi.org/10.34804/supra.2021092858 |
| Link: | https://doi.org/10.34804/supra.2021092858 |
| Export: | BibTex | RIS | EndNote |
Please find here information about the scholarly article describing the results derived from that data.
| Citation: |
|
| Link: | https://doi.org/10.1002/1099-1395(200101)14:1%3C11::AID-POC329%3E3.0.CO;2-7 |
| Export: | BibTex | RIS | EndNote |
Binding Isotherm Simulations
The plot depicts the binding isotherm simulation of a 1:1 interaction of Acridine red (0.0076045627376425855 M) and β-CD (0 — 0.015209125475285171 M).
Please sign in: customize the simulation by signing in to the SupraBank.