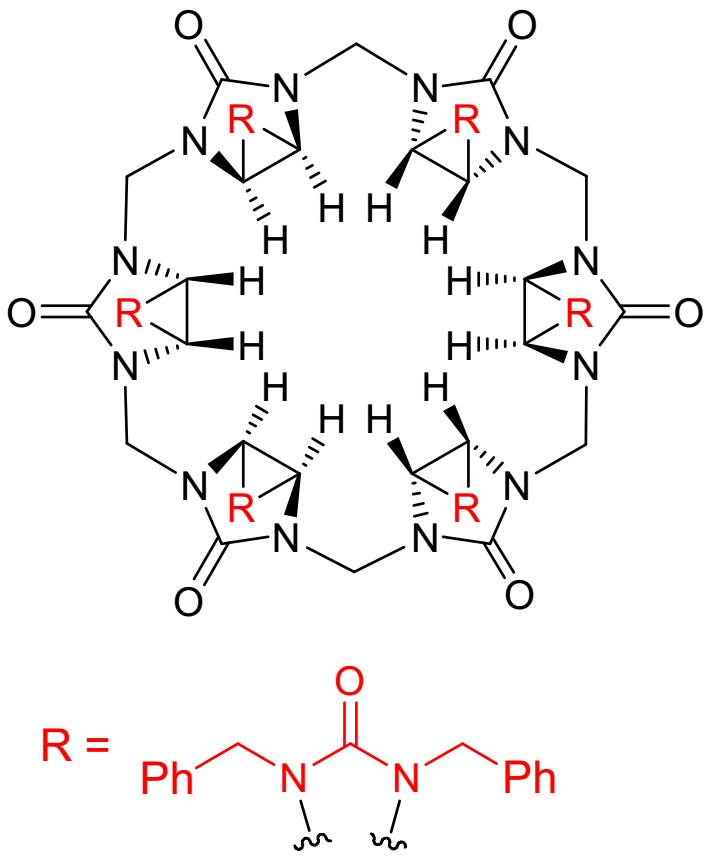
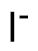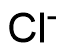Binding Properties
| 𝜈 | Molecule 1 : 1 Host | ||
| Ka = | 1.60⋅1010 | ± 6.40⋅108 | M-1 |
| Kd = | |||
| logKa = | |||
| T | 25.0 °C | ||
| Energy | kJ mol-1 | kcal mol-1 | |||
|---|---|---|---|---|---|
| ΔG | = | -58.25 | ± 0.1 | -13.92 | ± 0.02 |
| ΔH | = | -63.9 | ± 6.39 | -15.27 | ± 1.53 |
| -TΔS | = | 5.6 | 1.34 | ||
| J mol-1 K-1 | cal mol-1 K-1 | ||||
| ΔS | = | -18.8 | -4.5 | ||
These are the specifications of the determination of the experimental results.
| Detection Method: | Competitive | ||
| Assay Type: | Competitive Binding Assay | ||
| Technique: | Isothermal Titration Calorimetry | ||
| Molecule: | syringe | ||
| Partner: | cell | ||
| Indicator: | cell | ||
Detailed information about the solvation.
| Solvent System | Single Solvent |
| Solvent | chloroform |
Please find here information about the dataset this interaction is part of.
| Citation: |
V. Havel, V. Sindelar, SupraBank 2026, Anion Binding Inside a Bambus[6]uril Macrocycle in Chloroform (dataset). https://doi.org/10.34804/supra.20210928359 |
| Link: | https://doi.org/10.34804/supra.20210928359 |
| Export: | BibTex | RIS | EndNote |
Please find here information about the scholarly article describing the results derived from that data.
| Citation: |
V. Havel, V. Sindelar, ChemPlusChem 2015, 80, 1601–1606. |
| Link: | https://doi.org/10.1002/cplu.201500345 |
| Export: | BibTex | RIS | EndNote |
Binding Isotherm Simulations
The plot depicts the binding isotherm simulation of a 1:1 interaction of I- (1.25e-09 M) and Bn12BU (0 — 2.5e-09 M).
Please sign in: customize the simulation by signing in to the SupraBank.