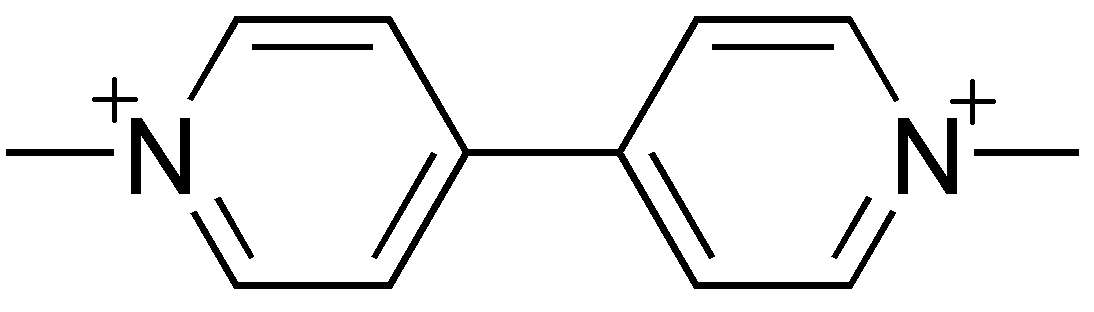
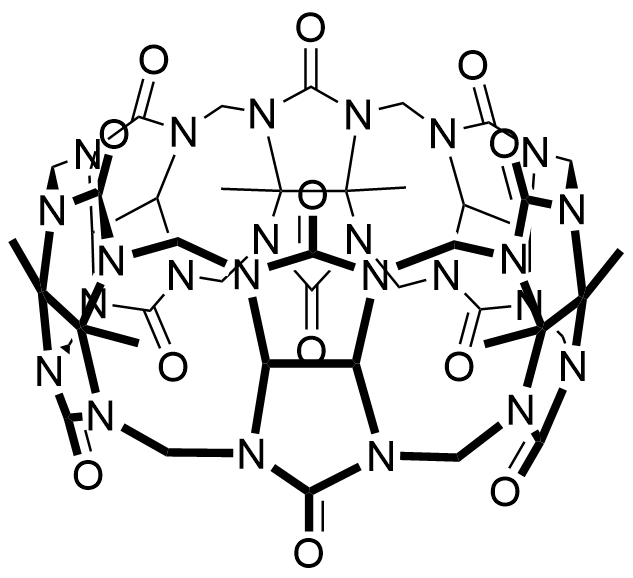
Binding Properties
| 𝜈 | Molecule 1 : 1 Host | ||
| Ka = | 1.97⋅105 | ± 1.30⋅104 | M-1 |
| Kd = | |||
| logKa = | |||
| T | 25.0 °C | ||
| Energy | kJ mol-1 | kcal mol-1 | |||
|---|---|---|---|---|---|
| ΔG | = | -30.22 | ± 0.16 | -7.22 | ± 0.04 |
These are the specifications of the determination of the experimental results.
| Detection Method: | Competitive | |||
| Assay Type: | Competitive Binding Assay | |||
| Technique: | Absorbance | |||
| 𝛌abs | = | 355.0 nm | ||
Detailed information about the solvation.
| Solvent System | Single Solvent |
| Solvent | water |
Please find here information about the dataset this interaction is part of.
| Citation: |
V. Sindelar, M. S. A. Khan, M. Necas, D. Heger, SupraBank 2026, Remarkable Salt Effect on Stability of Supramolecular Complex between Modified Cucurbit[6]uril and Methylviologen in Aqueous Media (dataset). https://doi.org/10.34804/supra.2021092833 |
| Link: | https://doi.org/10.34804/supra.2021092833 |
| Export: | BibTex | RIS | EndNote |
Please find here information about the scholarly article describing the results derived from that data.
| Citation: |
M. S. A. Khan, D. Heger, M. Necas, V. Sindelar, J. Phys. Chem. B 2009, 113, 11054–11057. |
| Link: | https://doi.org/10.1021/jp9059906 |
| Export: | BibTex | RIS | EndNote | |
Binding Isotherm Simulations
The plot depicts the binding isotherm simulation of a 1:1 interaction of paraquat ion (0.0001015228426395939 M) and Me6CB6 (0 — 0.0002030456852791878 M).
Please sign in: customize the simulation by signing in to the SupraBank.