Binding Properties
| 𝜈 | Molecule 1 : 1 Host | ||
| Ka = | 9800.0 | ± 950.0 | M-1 |
| Kd = | |||
| logKa = | |||
| T | 25.0 °C | ||
| Energy | kJ mol-1 | kcal mol-1 | |||
|---|---|---|---|---|---|
| ΔG | = | -22.78 | ± 0.24 | -5.44 | ± 0.06 |
These are the specifications of the determination of the experimental results.
| Detection Method: | Direct | ||
| Assay Type: | Direct Binding Assay | ||
| Technique: | Nuclear Magnetic Resonance | ||
| Nucleus | H-1 | ||
Detailed information about the solvation.
| Solvent System | Buffer System | 20 mM deuterated phosphate pD-7.4 |
| Solvents | Deuterium Oxide | |
| Additives | Disodium deuter... | 15.1 mM |
| Sodium dideuter... | 4.9 mM | |
| Source of Concentration | estimated | |
| Total concentration | 20.0 mM | |
| pH | 7.4 |
Please find here information about the dataset this interaction is part of.
| Citation: |
B. Zhang, L. Isaacs, V. Sindelar, L. Gilberg, P. Y. Zavalij, SupraBank 2026, Acyclic cucurbit[n]uril-type molecular containers: influence of glycoluril oligomer length on their function as solubilizing agents (dataset). https://doi.org/10.34804/supra.2021092839 |
| Link: | https://doi.org/10.34804/supra.2021092839 |
| Export: | BibTex | RIS | EndNote |
Please find here information about the scholarly article describing the results derived from that data.
| Citation: |
L. Gilberg, B. Zhang, P. Y. Zavalij, V. Sindelar, L. Isaacs, Org. Biomol. Chem. 2015, 13, 4041–4050. |
| Link: | https://doi.org/10.1039/C5OB00184F |
| Export: | BibTex | RIS | EndNote | |
Binding Isotherm Simulations
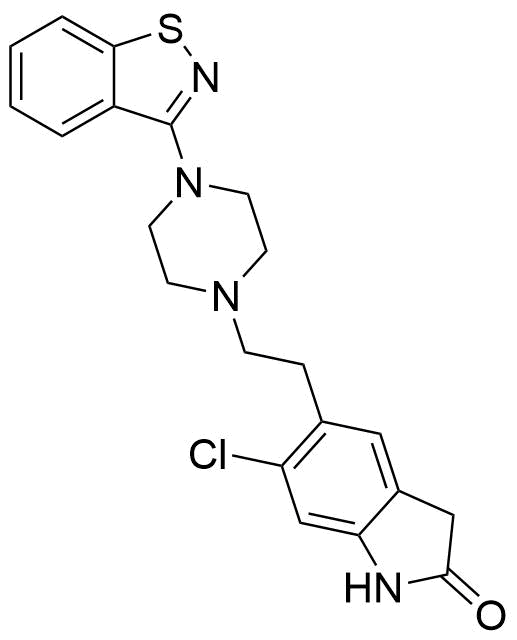
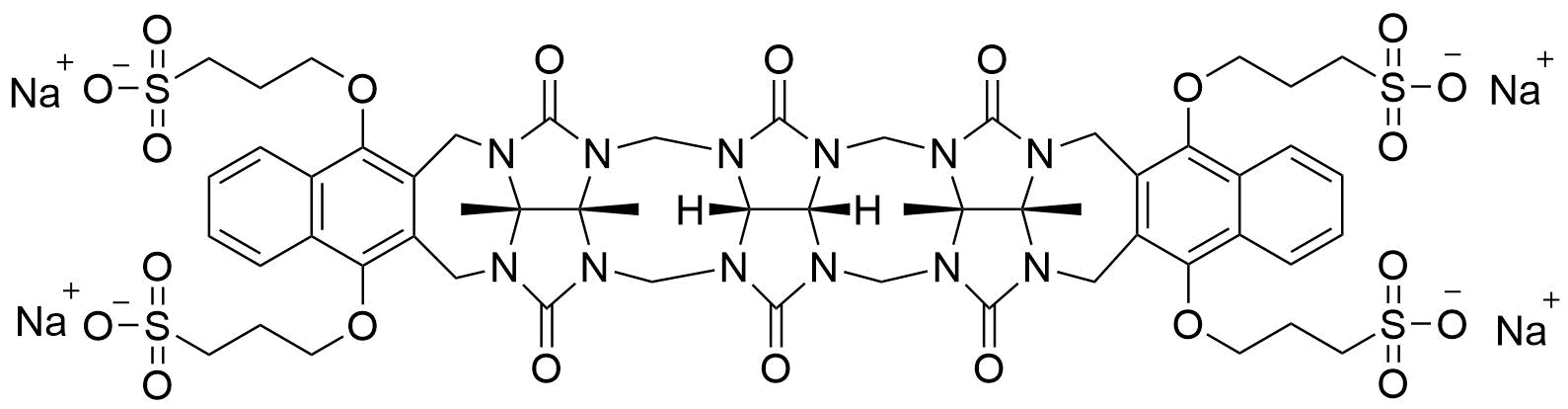
The plot depicts the binding isotherm simulation of a 1:1 interaction of Ziprasidone (0.0020408163265306124 M) and Dimetoxinaphtaleneglycoluril trimer (0 — 0.004081632653061225 M).
Please sign in: customize the simulation by signing in to the SupraBank.