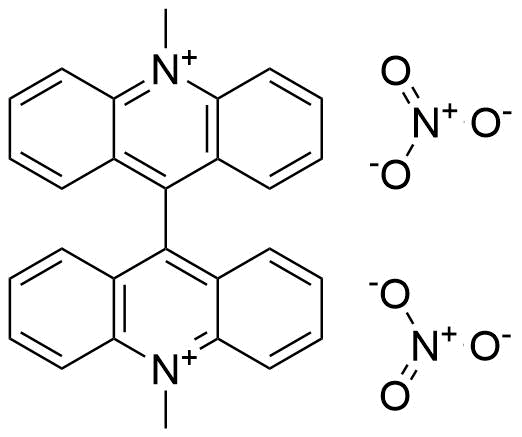
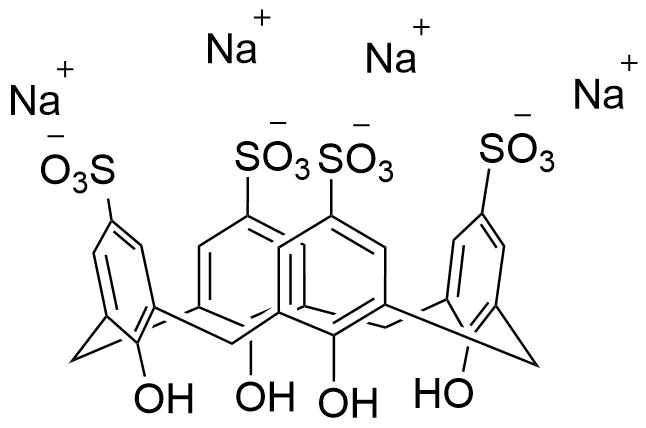
Binding Properties
| 𝜈 | Molecule 1 : 1 Host | ||
| Ka = | 1.60⋅106 | ± 1.00⋅105 | M-1 |
| Kd = | |||
| logKa = | |||
| T | 37.0 °C | ||
| Energy | kJ mol-1 | kcal mol-1 | |||
|---|---|---|---|---|---|
| ΔG | = | -36.84 | ± 0.16 | -8.8 | ± 0.04 |
These are the specifications of the determination of the experimental results.
| Detection Method: | Direct | |||
| Assay Type: | Direct Binding Assay | |||
| Technique: | Fluorescence | |||
| 𝛌ex | = | 368.0 nm | ||
| 𝛌em | = | 510.0 nm | ||
Detailed information about the solvation.
| Solvent System | Buffer System | 10 mM phosphate pH-2.0 |
| Solvents | water | |
| Additives | Phosphoric acid | 10.0 mM |
| Sodium hydroxide | ||
| Source of Concentration | estimated | |
| Total concentration | 10.0 mM | |
| pH | 2.0 |
Please find here information about the dataset this interaction is part of.
| Citation: |
D. Guo, Z. Zheng, W. Geng, H. Li, SupraBank 2026, Sensitive fluorescence detection of saliva pepsin by a supramolecular tandem assay enables the diagnosis of gastroesophageal reflux disease (dataset). https://doi.org/10.34804/supra.20210928256 |
| Link: | https://doi.org/10.34804/supra.20210928256 |
| Export: | BibTex | RIS | EndNote |
Please find here information about the scholarly article describing the results derived from that data.
| Citation: |
Z. Zheng, W.-C. Geng, H.-B. Li, D.-S. Guo, Supramolecular Chemistry 2020, 1–8. |
| Link: | https://doi.org/10.1080/10610278.2020.1857762 |
| Export: | BibTex | RIS | EndNote |
Binding Isotherm Simulations
The plot depicts the binding isotherm simulation of a 1:1 interaction of Lucigenin (1.25e-05 M) and sCx4 sodium salt (0 — 2.5e-05 M).
Please sign in: customize the simulation by signing in to the SupraBank.