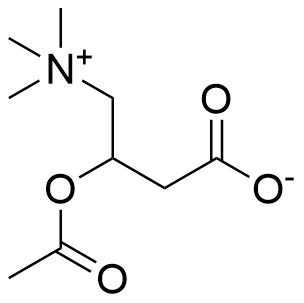
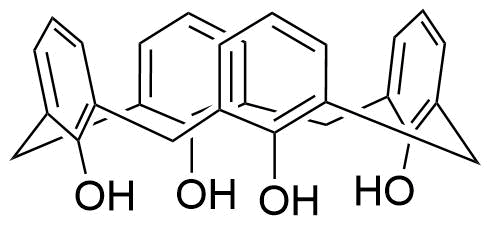
Binding Properties
| 𝜈 | Molecule 1 : 1 Host | ||
| Ka = | 2900.0 | ± 500.0 | M-1 |
| Kd = | |||
| logKa = | |||
| T | 25.0 °C | ||
| Energy | kJ mol-1 | kcal mol-1 | |||
|---|---|---|---|---|---|
| ΔG | = | -19.76 | ± 0.43 | -4.72 | ± 0.1 |
These are the specifications of the determination of the experimental results.
| Detection Method: | Competitive | |||
| Assay Type: | Competitive Binding Assay | |||
| Technique: | Fluorescence | |||
| 𝛌ex | = | 365.0 nm | ||
Detailed information about the solvation.
| Solvent System | Complex Mixture |
| Solvents | Deuterium Oxide |
| Additives | Deuterium chloride |
| pH | 2.4 |
Please find here information about the dataset this interaction is part of.
| Citation: |
H. Bakirci, W. M. Nau, SupraBank 2026, Fluorescence Regeneration as a Signaling Principle for Choline and Carnitine Binding: A Refined Supramolecular Sensor System Based on a Fluorescent Azoalkane (dataset). https://doi.org/10.34804/supra.20210928371 |
| Link: | https://doi.org/10.34804/supra.20210928371 |
| Export: | BibTex | RIS | EndNote |
Please find here information about the scholarly article describing the results derived from that data.
| Citation: |
H. Bakirci, W. M. Nau, Adv. Funct. Mater. 2006, 16, 237–242. |
| Link: | https://doi.org/10.1002/adfm.200500219 |
| Export: | BibTex | RIS | EndNote |
Binding Isotherm Simulations
The plot depicts the binding isotherm simulation of a 1:1 interaction of Acetyl-D/L-Carnitine (0.006896551724137931 M) and Cx4 (0 — 0.013793103448275862 M).
Please sign in: customize the simulation by signing in to the SupraBank.