Binding Properties
| 𝜈 | Molecule 1 : 1 Host | ||
| Ka = | 1.40⋅104 | ± 5000.0 | M-1 |
| Kd = | |||
| logKa = | |||
| T | 25.0 °C | ||
| Energy | kJ mol-1 | kcal mol-1 | |||
|---|---|---|---|---|---|
| ΔG | = | -23.67 | ± 0.93 | -5.66 | ± 0.22 |
| ΔH | = | -53.6 | ± 1.5 | -12.81 | ± 0.36 |
| -TΔS | = | 26.8 | ± 2.0 | 6.41 | ± 0.48 |
| J mol-1 K-1 | cal mol-1 K-1 | ||||
| ΔS | = | -89.9 | ± 6.7 | -21.5 | ± 1.6 |
These are the specifications of the determination of the experimental results.
| Detection Method: | Direct | ||
| Assay Type: | Associative Binding Assay | ||
| Technique: | Isothermal Titration Calorimetry | ||
| Instrument: | MicroCal VP-ITC | ||
| VCell | = | 1400.0 𝜇L | |
| VSyringe | = | 350.0 𝜇L | |
| cmolecule | = | 1200.0 𝜇M syringe | |
| cpartner | = | 100.0 𝜇M cell | |
| ccofactor | = | 100.0 𝜇M cell | |
| Ninjection | = | 30 | |
| Vinjection | = | 10.0 𝜇L | |
| Vinit | = | 2.0 𝜇L | |
Detailed information about the solvation.
| Solvent System | Single Solvent |
| Solvent | water |
| pH | 7.0 |
Please find here information about the dataset this interaction is part of.
| Citation: |
O. Scherman, F. Biedermann, J. del Barrio, E. Appel, D. Hoogland, M. D. Driscoll, S. Hay, D. J. Wales, SupraBank 2026, Decoupled Associative and Dissociative Processes in Strong yet Highly Dynamic Host–Guest Complexes (dataset). https://doi.org/10.34804/supra.20220701456 |
| Link: | https://doi.org/10.34804/supra.20220701456 |
| Export: | BibTex | RIS | EndNote |
Please find here information about the scholarly article describing the results derived from that data.
| Citation: |
E. A. Appel, F. Biedermann, D. Hoogland, J. del Barrio, M. D. Driscoll, S. Hay, D. J. Wales, O. A. Scherman, J. Am. Chem. Soc. 2017, 139, 12985–12993. |
| Link: | https://doi.org/10.1021/jacs.7b04821 |
| Export: | BibTex | RIS | EndNote | |
Binding Isotherm Simulations
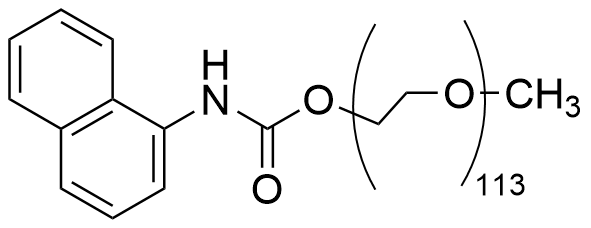
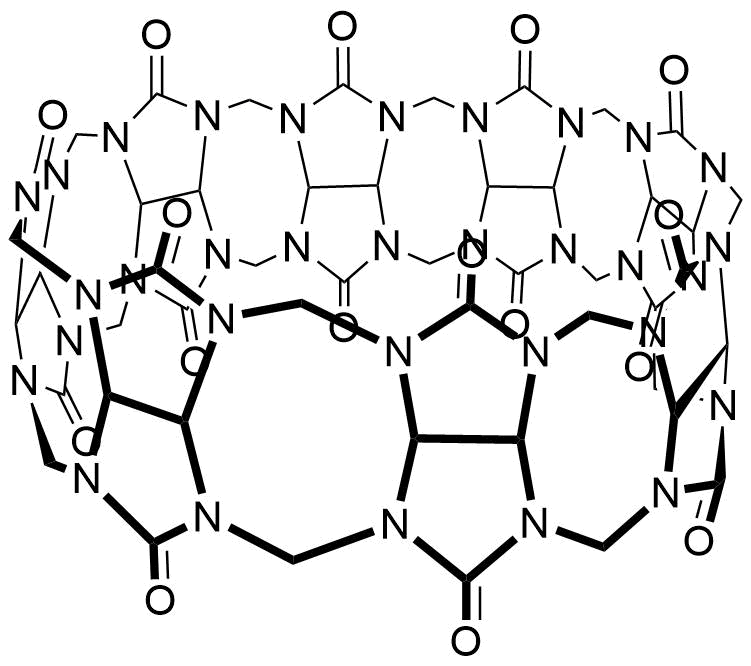
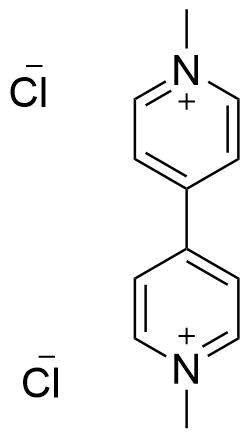
The plot depicts the binding isotherm simulation of a 1:1 interaction of 1Np-5kPEGOMe (0.0014285714285714286 M) and CB8 (0 — 0.002857142857142857 M).
Please sign in: customize the simulation by signing in to the SupraBank.