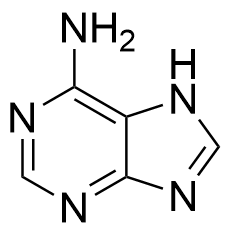
Binding Properties
| 𝜈 | Molecule 1 : 1 Host | ||
| Ka = | 2060.0 | ± 120.0 | M-1 |
| Kd = | |||
| logKa = | |||
| T | 25.0 °C | ||
| Energy | kJ mol-1 | kcal mol-1 | |||
|---|---|---|---|---|---|
| ΔG | = | -18.92 | ± 0.14 | -4.52 | ± 0.03 |
- Comment
- Δδ was estimated graphically
These are the specifications of the determination of the experimental results.
| Detection Method: | Direct | ||
| Assay Type: | Direct Binding Assay | ||
| Technique: | Nuclear Magnetic Resonance | ||
| Nucleus | H-1 | ||
| = | 7.58 ppm | ||
| = | 7.76 ppm | ||
| = | 0.18 ppm | ||
| δbound⁄δfree | = | 1.02 | |
Detailed information about the solvation.
| Solvent System | Single Solvent |
| Solvent | Deuterium Oxide |
Please find here information about the dataset this interaction is part of.
| Citation: |
S. Zhuang, G. Wu, J. Sun, J. Zhang, J. Xing, Y. Wu, H. Wang, Z. Li, D. Zhang, SupraBank 2025, Water-soluble hexakis-imidazolium cages (dataset). https://doi.org/10.34804/supra.20250408599 |
| Link: | https://doi.org/10.34804/supra.20250408599 |
| Export: | BibTex | RIS | EndNote |
Please find here information about the scholarly article describing the results derived from that data.
| Citation: |
S.-Y. Zhuang, G. Wu, J.-D. Sun, J. Zhang, J. Xing, Y. Wu, H. Wang, Z.-T. Li, D.-W. Zhang, Org. Chem. Front. 2024, 11, 414–421. |
| Link: | https://doi.org/10.1039/d3qo01780j |
| Export: | BibTex | RIS | EndNote | |
Binding Isotherm Simulations
The plot depicts the binding isotherm simulation of a 1:1 interaction of Adenine (0.009708737864077669 M) and p-HiCage (0 — 0.019417475728155338 M).
Please sign in: customize the simulation by signing in to the SupraBank.