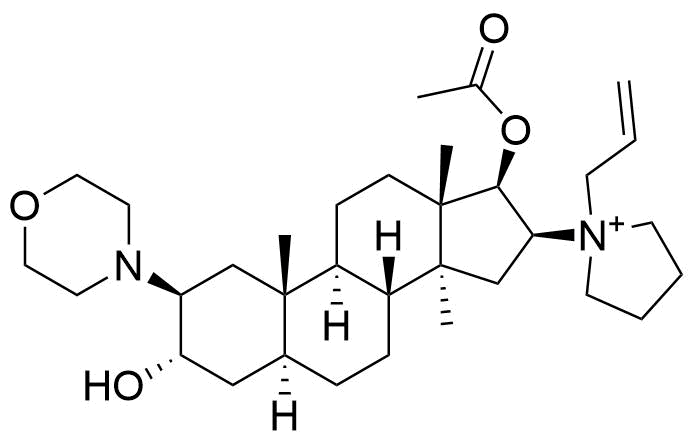
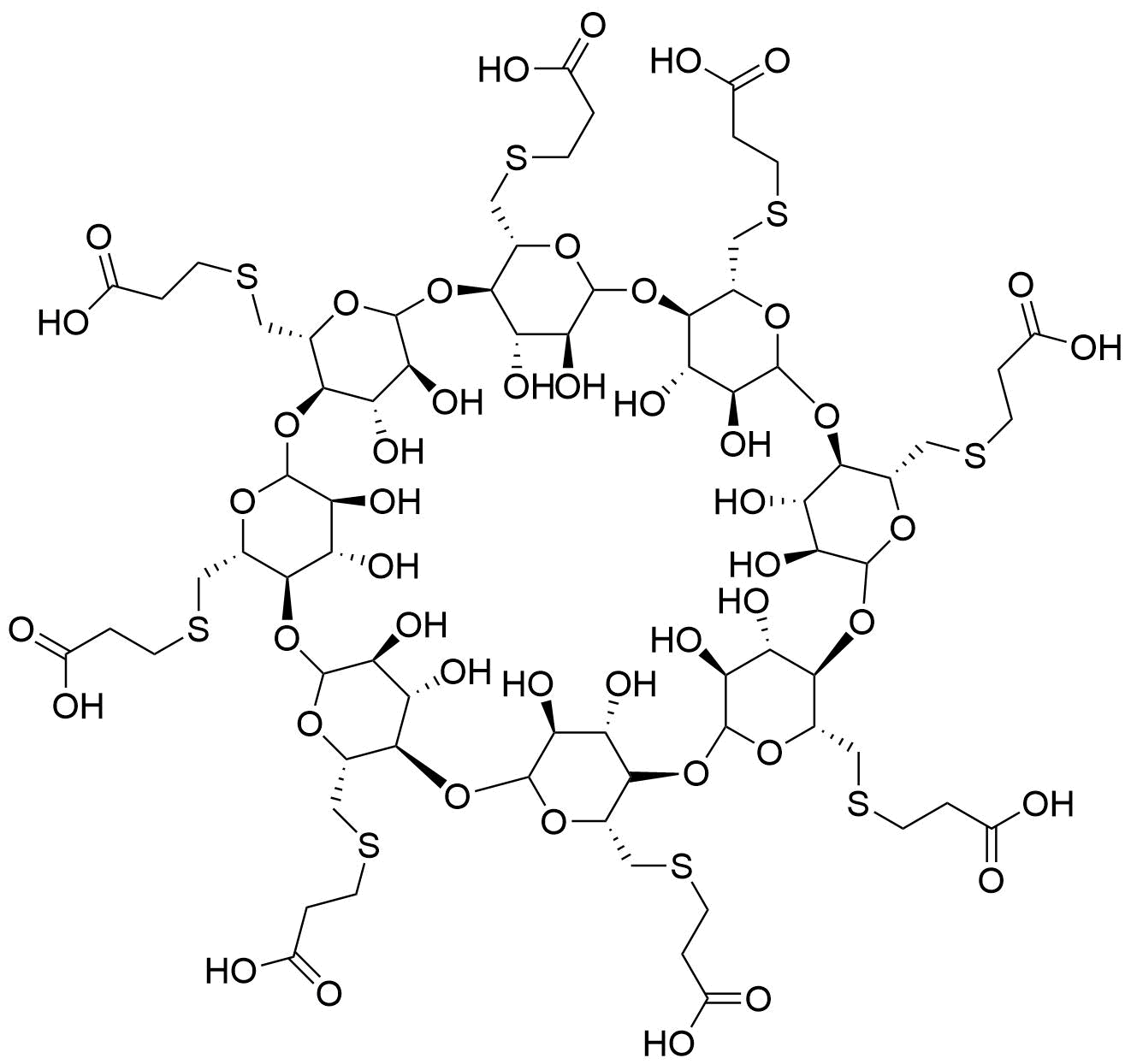
Binding Properties
| 𝜈 | Molecule 1 : 1 Host | ||
| Ka = | 1.05⋅107 | ± 1.60⋅106 | M-1 |
| Kd = | |||
| logKa = | |||
| T | 25.0 °C | ||
| Energy | kJ mol-1 | kcal mol-1 | |||
|---|---|---|---|---|---|
| ΔG | = | -40.08 | ± 0.38 | -9.58 | ± 0.09 |
| ΔH | = | -27.86 | ± 0.25 | -6.66 | ± 0.06 |
| -TΔS | = | -12.22 | -2.92 | ||
| J mol-1 K-1 | cal mol-1 K-1 | ||||
| ΔS | = | 41.0 | 9.8 | ||
These are the specifications of the determination of the experimental results.
| Detection Method: | Direct | ||
| Assay Type: | Direct Binding Assay | ||
| Technique: | Isothermal Titration Calorimetry | ||
| Molecule: | syringe | ||
| Partner: | cell | ||
Detailed information about the solvation.
| Solvent System | Buffer System | 50 mM phosphate pH-7 |
| Solvents | water | |
| Source of Concentration | ||
| Total concentration | 50.0 mM | |
| pH | 7.0 |
Please find here information about the dataset this interaction is part of.
| Citation: |
A. Bom, M. Bradley, K. Cameron, J. K. Clark, J. van Egmond, H. Feilden, E. J. MacLean, A. W. Muir, R. Palin, D. C. Rees, M. Zhang, SupraBank 2026, A Novel Concept of Reversing Neuromuscular Block: Chemical Encapsulation of Rocuronium Bromide by a Cyclodextrin-Based Synthetic Host (dataset). https://doi.org/10.34804/supra.20211020406 |
| Link: | https://doi.org/10.34804/supra.20211020406 |
| Export: | BibTex | RIS | EndNote |
Please find here information about the scholarly article describing the results derived from that data.
| Citation: |
A. Bom, M. Bradley, K. Cameron, J. K. Clark, J. van Egmond, H. Feilden, E. J. MacLean, A. W. Muir, R. Palin, D. C. Rees, et al., Angew. Chem. Int. Ed. 2002, 41, 265. |
| Link: | https://doi.org/10.1002/1521-3773(20020118)41:2%3C265::AID-ANIE265%3E3.0.CO;2-Q |
| Export: | BibTex | RIS | EndNote |
Binding Isotherm Simulations
The plot depicts the binding isotherm simulation of a 1:1 interaction of Rocuronium (1.9047619047619047e-06 M) and Sugammadex (0 — 3.8095238095238094e-06 M).
Please sign in: customize the simulation by signing in to the SupraBank.