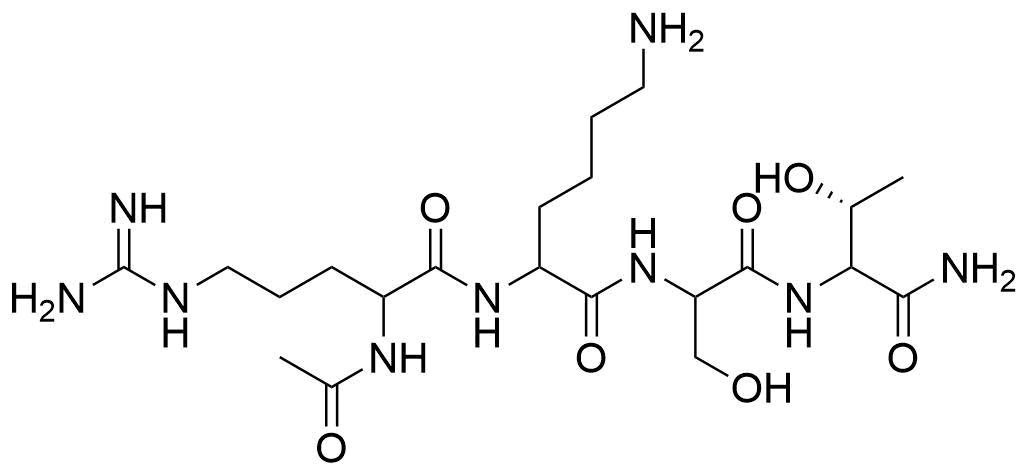
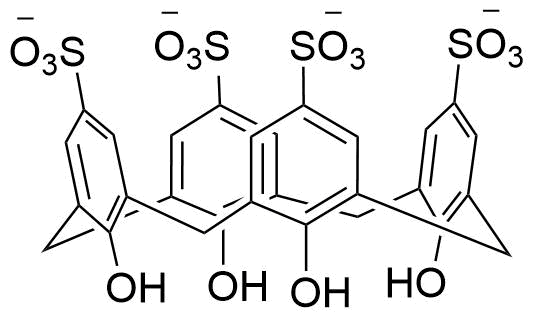
Binding Properties
| 𝜈 | Molecule 1 : 1 Host | ||
| Ka = | 1.52⋅104 | ± 4000.0 | M-1 |
| Kd = | |||
| logKa = | |||
| T | 25.0 °C | ||
| Energy | kJ mol-1 | kcal mol-1 | |||
|---|---|---|---|---|---|
| ΔG | = | -23.87 | ± 0.67 | -5.71 | ± 0.16 |
These are the specifications of the determination of the experimental results.
| Detection Method: | Direct | ||
| Assay Type: | Direct Binding Assay | ||
| Technique: | Nuclear Magnetic Resonance | ||
Detailed information about the solvation.
| Solvent System | Buffer System | 40 mM deuterated phosphate pD-7.4 |
| Solvents | Deuterium Oxide | 100.0 % |
| Additives | Disodium hydrog... | 30.2 mM |
| Sodium dihydrog... | 9.8 mM | |
| Source of Concentration | ||
| Total concentration | 40.0 mM | |
| pH | 7.4 |
Please find here information about the dataset this interaction is part of.
| Citation: |
K. D. Daze, F. Hof, C. S. Beshara, C. E. Jones, B. J. Lilgert, SupraBank 2026, A Simple Calixarene Recognizes Post-translationally Methylated Lysine (dataset). https://doi.org/10.34804/supra.2021092818 |
| Link: | https://doi.org/10.34804/supra.2021092818 |
| Export: | BibTex | RIS | EndNote |
Please find here information about the scholarly article describing the results derived from that data.
| Citation: |
C. S. Beshara, C. E. Jones, K. D. Daze, B. J. Lilgert, F. Hof, Chem. Eur. J. of Chem. Bio. 2009, 11, 63–66. |
| Link: | https://doi.org/10.1002/cbic.200900633 |
| Export: | BibTex | RIS | EndNote |
Binding Isotherm Simulations
The plot depicts the binding isotherm simulation of a 1:1 interaction of RKST (0.0013157894736842105 M) and sCx4 (0 — 0.002631578947368421 M).
Please sign in: customize the simulation by signing in to the SupraBank.