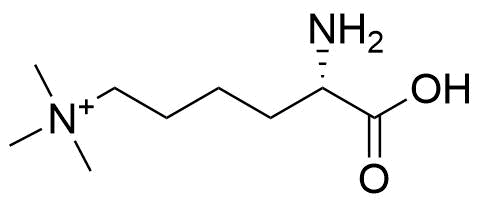
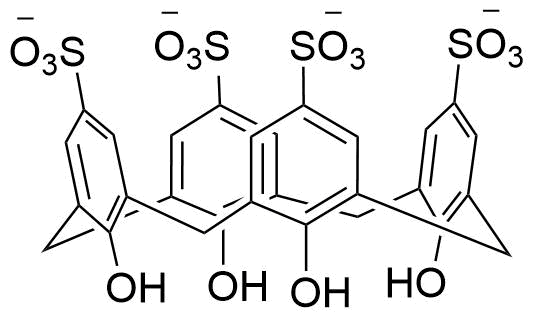
Binding Properties
| 𝜈 | Molecule 1 : 1 Host | ||
| Ka = | 1.23⋅104 | ± 550.0 | M-1 |
| Kd = | |||
| logKa = | |||
| T | 37.0 °C | ||
| Energy | kJ mol-1 | kcal mol-1 | |||
|---|---|---|---|---|---|
| ΔG | = | -24.28 | ± 0.11 | -5.8 | ± 0.03 |
| ΔH | = | -20.4 | -4.88 | ||
| -TΔS | = | -3.9 | -0.93 | ||
| J mol-1 K-1 | cal mol-1 K-1 | ||||
| ΔS | = | 12.6 | 3.0 | ||
These are the specifications of the determination of the experimental results.
| Detection Method: | Direct | ||
| Assay Type: | Direct Binding Assay | ||
| Technique: | Isothermal Titration Calorimetry | ||
| Molecule: | syringe | ||
| Partner: | syringe | ||
Detailed information about the solvation.
| Solvent System | Buffer System | 40 mM PBS pH-7.4 |
| Solvents | water | |
| Additives | sodium chloride | 137.0 mM |
| Disodium hydrog... | 30.2 mM | |
| Sodium dihydrog... | 9.8 mM | |
| potassium chloride | 2.7 mM | |
| Source of Concentration | ||
| Total concentration | 40.0 mM | |
| pH | 7.4 |
Please find here information about the dataset this interaction is part of.
| Citation: |
K. D. Daze, F. Hof, C. S. Beshara, C. E. Jones, B. J. Lilgert, SupraBank 2026, Determining the effects of salt, buffer, and temperature on the complexation of methylated ammonium ions and methyllysines by sulfonated calixarenes (dataset). https://doi.org/10.34804/supra.2021092877 |
| Link: | https://doi.org/10.34804/supra.2021092877 |
| Export: | BibTex | RIS | EndNote |
Please find here information about the scholarly article describing the results derived from that data.
| Citation: |
K. D. Daze, C. E. Jones, B. J. Lilgert, C. S. Beshara, F. Hof, Can. J. Chem. 2013, 91, 1072–1076. |
| Link: | https://doi.org/10.1139/cjc-2013-0186 |
| Export: | BibTex | RIS | EndNote |
Binding Isotherm Simulations
The plot depicts the binding isotherm simulation of a 1:1 interaction of Trimethyllysine (0.0016260162601626016 M) and sCx4 (0 — 0.0032520325203252032 M).
Please sign in: customize the simulation by signing in to the SupraBank.