Binding Properties
| 𝜈 | Molecule 1 : 1 Host | ||
| Ka = | 1500.0 | ± 600.0 | M-1 |
| Kd = | |||
| logKa = | |||
| T | 25.0 °C | ||
| Energy | kJ mol-1 | kcal mol-1 | |||
|---|---|---|---|---|---|
| ΔG | = | -18.13 | ± 1.05 | -4.33 | ± 0.25 |
| Detection Method: | Direct | ||
| Assay Type: | Direct Binding Assay | ||
| Technique: | Nuclear Magnetic Resonance | ||
| Solvent System | Buffer System | 10 mM phosphate pH-8.0 |
| Solvents | water | |
| Additives | Sodium dihydrog... | |
| Disodium hydrog... | ||
| Source of Concentration | ||
| Total concentration | 10.0 mM | |
| pH | 8.0 |
| Citation: |
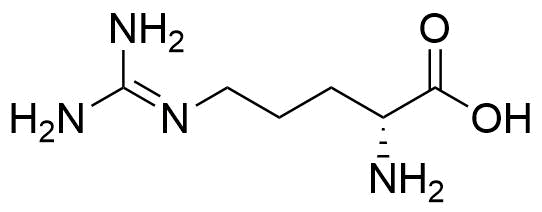
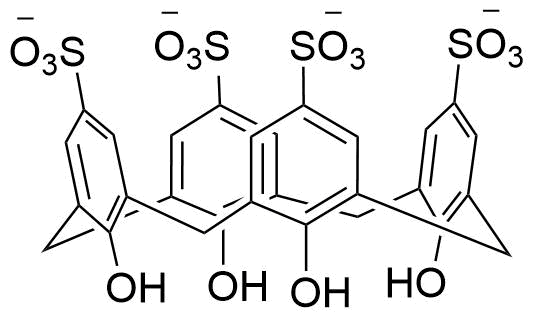
F. Perret, J. Morel, N. Morel-Desrosiers, N. Douteau-Guével, A. W. Coleman, SupraBank 2026, Binding of dipeptides and tripeptides containing lysine or arginine by p-sulfonatocalixarenes in water: NMR and microcalorimetric studiesElectronic supplementary information (ESI) available: chemical shifts experienced by different protons of KK, RR, KKK or RRR in the presence of increasing amounts of 14 or 16; heat effects observed upon titration of 14, 16 or 18 by KK, RR, KKK or RRR; COSY and ROESY 2D 1H NMR spectra for 14 complexed with KK or RR. See http://www.rsc.org/suppdata/p2/b1/b109553f/ (dataset). https://doi.org/10.34804/supra.20210928255 |
| Link: | https://doi.org/10.34804/supra.20210928255 |
| Export: | BibTex | RIS | EndNote |
| Citation: |
N. Douteau-Guével, F. Perret, A. W. Coleman, J.-P. Morel, N. Morel-Desrosiers, J. Chem. Soc., Perkin Trans. 2 2002, 524–532. |
| Link: | https://doi.org/10.1039/B109553F |
| Export: | BibTex | RIS | EndNote | |
Binding Isotherm Simulations
The plot depicts the binding isotherm simulation of a 1:1 interaction of L-Arg (0.013333333333333334 M) and sCx4 (0 — 0.02666666666666667 M).