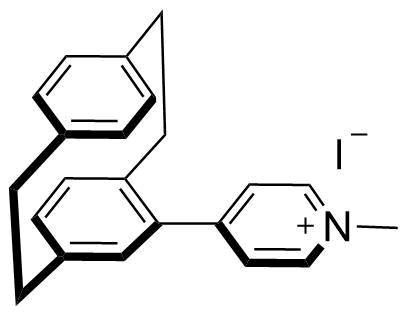
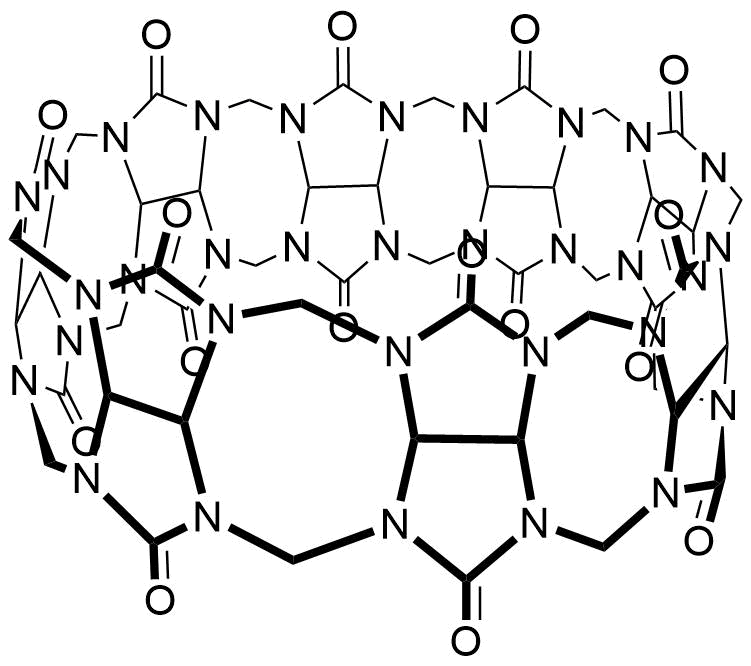
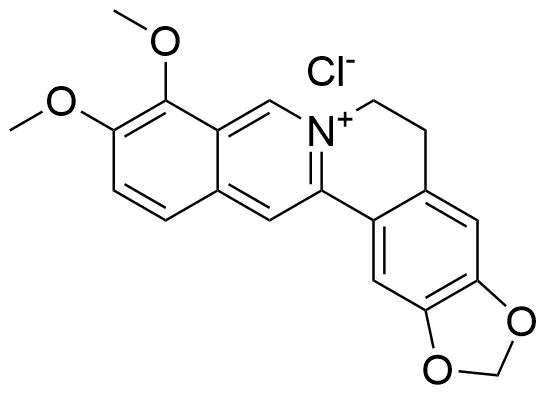
Binding Properties
| 𝜈 | Molecule 1 : 1 Host | ||
| Ka = | 7.55⋅1010 | ± 1.68⋅109 | M-1 |
| Kd = | |||
| logKa = | |||
| T | 25.0 °C | ||
| Energy | kJ mol-1 | kcal mol-1 | |||
|---|---|---|---|---|---|
| ΔG | = | -62.09 | ± 0.56 | -14.84 | ± 0.13 |
These are the specifications of the determination of the experimental results.
| Detection Method: | Competitive | |||
| Assay Type: | Competitive Binding Assay | |||
| Technique: | Fluorescence | |||
| 𝛌ex | = | 342.0 nm | ||
| 𝛌em | = | 542.0 nm | ||
| Ibound⁄Ifree | = | 6.8 | ||
Detailed information about the solvation.
| Solvent System | Buffer System | 1X PBS pH-7.4 |
| Solvents | water | |
| Additives | sodium chloride | 137.0 mM |
| Disodium hydrog... | 10.0 mM | |
| potassium chloride | 2.7 mM | |
| Potassium dihyd... | 1.8 mM | |
| Source of Concentration | real | |
| Total concentration | 151.5 mM | |
| pH | 7.4 |
Please find here information about the dataset this interaction is part of.
| Citation: |
F. Biedermann, S. Sinn, E. Spuling, S. Braese, SupraBank 2026, Rational design and implementation of a cucurbit[8]uril-based indicator-displacement assay for application in blood serum (dataset). https://doi.org/10.34804/supra.2021092883 |
| Link: | https://doi.org/10.34804/supra.2021092883 |
| Export: | BibTex | RIS | EndNote |
Please find here information about the scholarly article describing the results derived from that data.
| Citation: |
S. Sinn, E. Spuling, S. Bräse, F. Biedermann, Chem. Sci. 2019, 10, 6584–6593. |
| Link: | https://doi.org/10.1039/C9SC00705A |
| Export: | BibTex | RIS | EndNote | |
Binding Isotherm Simulations
The plot depicts the binding isotherm simulation of a 1:1 interaction of MPCP (2.649006622516556e-10 M) and CB8 (0 — 5.298013245033112e-10 M).
Please sign in: customize the simulation by signing in to the SupraBank.