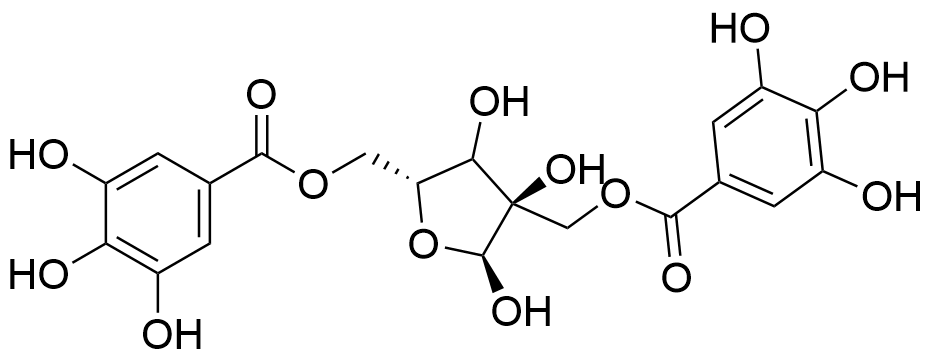
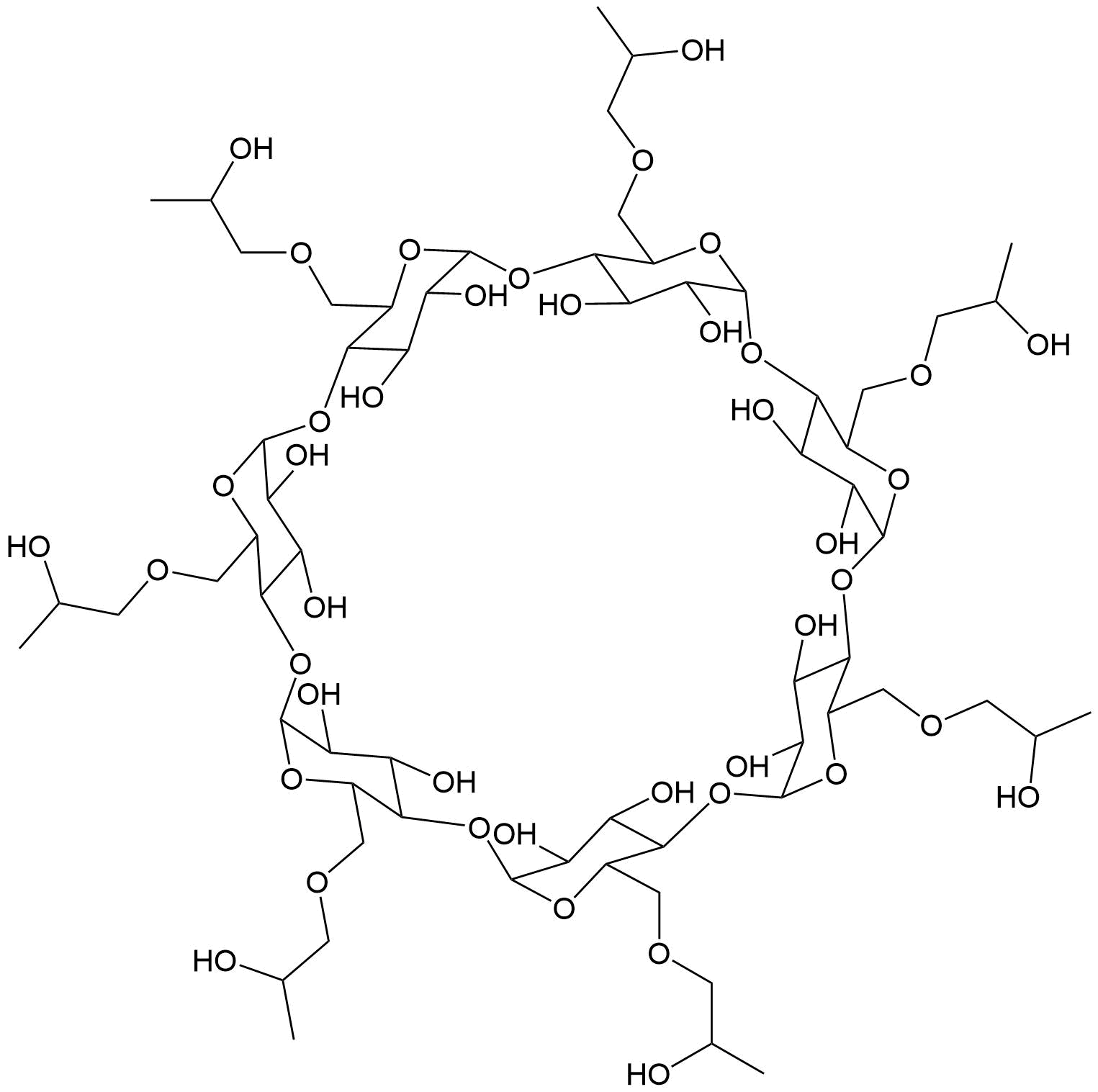
Binding Properties
| 𝜈 | Molecule 1 : 1 Host | ||
| Ka = | 1.61⋅104 | ± 2200.0 | M-1 |
| Kd = | |||
| logKa = | |||
| T | 25.0 °C | ||
| Energy | kJ mol-1 | kcal mol-1 | |||
|---|---|---|---|---|---|
| ΔG | = | -24.01 | ± 0.34 | -5.74 | ± 0.08 |
| ΔH | = | -0.38 | ± 0.04 | -0.09 | ± 0.01 |
| -TΔS | = | -25.6 | ± 3.7 | -6.12 | ± 0.88 |
| J mol-1 K-1 | cal mol-1 K-1 | ||||
| ΔS | = | 85.9 | ± 12.4 | 20.5 | ± 3.0 |
- Comment
- One-site binding model (HPβCD1:HAM1)
| Detection Method: | Direct | ||
| Assay Type: | Direct Binding Assay | ||
| Technique: | Isothermal Titration Calorimetry | ||
| Instrument: | MicroCal VP-ITC | ||
| VCell | = | 1400.0 𝜇L | |
| VSyringe | = | 350.0 𝜇L | |
| Molecule: | syringe | ||
| Partner: | cell | ||
| Solvent System | Single Solvent |
| Solvent | water |
| Citation: |
G. Brackman, J. Lenoir, L. De Meyer, T. De Beer, A. Concheiro, T. Coenye, M. J. Garcia‐Fernandez, J. Remon, C. Alvarez‐Lorenzo, SupraBank 2025, Dressings Loaded with Cyclodextrin–Hamamelitannin Complexes Increase Staphylococcus aureus Susceptibility Toward Antibiotics Both in Single as well as in Mixed Biofilm Communities (dataset). https://doi.org/10.34804/supra.20240328543 |
| Link: | https://doi.org/10.34804/supra.20240328543 |
| Export: | BibTex | RIS | EndNote |
| Citation: |
G. Brackman, M. J. Garcia-Fernandez, J. Lenoir, L. De Meyer, J.-P. Remon, T. De Beer, A. Concheiro, C. Alvarez-Lorenzo, T. Coenye, Macromol. Biosci. 2016, 16, 859–869. |
| Link: | https://doi.org/10.1002/mabi.201500437 |
| Export: | BibTex | RIS | EndNote |
Binding Isotherm Simulations
The plot depicts the binding isotherm simulation of a 1:1 interaction of Hamamelitannin (0.0012422360248447205 M) and heptakis-O-(2-hydroxypropyl)-β Cyclodextrin (0 — 0.002484472049689441 M).