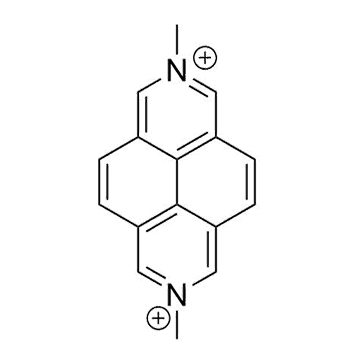
Binding Properties
| 𝜈 | Molecule 1 : 1 Host | ||
| Ka = | 2.69⋅109 | M-1 | |
| Kd = | |||
| logKa = | |||
| T | 25.0 °C | ||
| kin= | 24000000.0 | ± 5000000.0 | M-1s-1 |
| kout= | 0.00891530460624071 | ± 0.0018 | s-1 |
| Energy | kJ mol-1 | kcal mol-1 | |||
|---|---|---|---|---|---|
| ΔG | = | -53.83 | -12.87 |
These are the specifications of the determination of the experimental results.
| Detection Method: | Direct | |||
| Assay Type: | Direct Binding Assay | |||
| Technique: | Fluorescence | |||
| 𝛌ex | = | 339.0 nm | ||
| 𝛌em | = | 454.0 nm | ||
Detailed information about the solvation.
| Solvent System | Single Solvent |
| Solvent | water |
| pH | 7.0 |
Please find here information about the dataset this interaction is part of.
| Citation: |
Z. Miskolczy, L. Biczok, L. M. Grimm, F. Biedermann, M. Megyesi, S. Sinn, S. Braese, A. Prabodh, SupraBank 2026, Teaching indicators to unravel the kinetic features of host–guest inclusion complexes (dataset). https://doi.org/10.34804/supra.20220717459 |
| Link: | https://doi.org/10.34804/supra.20220717459 |
| Export: | BibTex | RIS | EndNote |
Please find here information about the scholarly article describing the results derived from that data.
| Citation: |
A. Prabodh, S. Sinn, L. Grimm, Z. Miskolczy, M. Megyesi, L. Biczók, S. Bräse, F. Biedermann, Chem. Commun. 2020, 56, 12327–12330. |
| Link: | https://doi.org/10.1039/D0CC03715J |
| Export: | BibTex | RIS | EndNote | |
Binding Isotherm Simulations
The plot depicts the binding isotherm simulation of a 1:1 interaction of MDAP (7.4294205052005945e-09 M) and CB7 (0 — 1.4858841010401189e-08 M).
Please sign in: customize the simulation by signing in to the SupraBank.