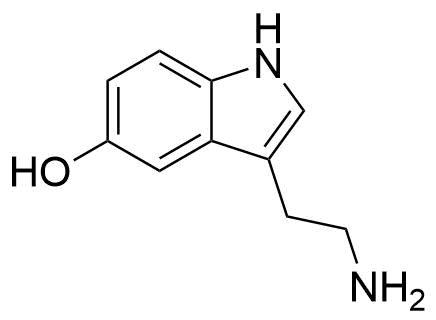
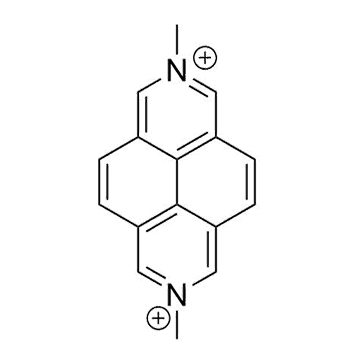
Binding Properties
| 𝜈 | Molecule 1 : 1 Host | ||
| Ka = | 6.85⋅104 | ± 1880.0 | M-1 |
| Kd = | |||
| logKa = | |||
| T | 25.0 °C | ||
| Energy | kJ mol-1 | kcal mol-1 | |||
|---|---|---|---|---|---|
| ΔG | = | -27.6 | ± 0.07 | -6.6 | ± 0.02 |
These are the specifications of the determination of the experimental results.
| Detection Method: | Direct | |||
| Assay Type: | Associative Binding Assay | |||
| Technique: | Fluorescence | |||
| 𝛌ex | = | 371.0 nm | ||
| 𝛌em | = | 424.0 nm | ||
Detailed information about the solvation.
| Solvent System | Buffer System | 1X PBS pH-7.0 |
| Solvents | water | |
| Additives | sodium chloride | 137.0 mM |
| Disodium hydrog... | 10.0 mM | |
| potassium chloride | 2.7 mM | |
| Potassium dihyd... | 1.8 mM | |
| Source of Concentration | ||
| Total concentration | 151.5 mM | |
| pH | 7.0 | |
| Suspension | The parnter/host is present as a suspension. |
Please find here information about the dataset this interaction is part of.
| Citation: |
L. M. Grimm, F. Biedermann, S. Sinn, M. Krstić, E. D'Este, I. Sonntag, E. A. Prasetyanto, T. Kuner, W. Wenzel, L. De Cola, SupraBank 2026, Fluorescent Nanozeolite Receptors for the Highly Selective and Sensitive Detection of Neurotransmitters in Water and Biofluids (dataset). https://doi.org/10.34804/supra.20220323419 |
| Link: | https://doi.org/10.34804/supra.20220323419 |
| Export: | BibTex | RIS | EndNote |
Please find here information about the scholarly article describing the results derived from that data.
| Citation: |
L. M. Grimm, S. Sinn, M. Krstić, E. D'Este, I. Sonntag, E. A. Prasetyanto, T. Kuner, W. Wenzel, L. De Cola, F. Biedermann, Adv. Mater. 2021, 33, 2104614. |
| Link: | https://doi.org/10.1002/adma.202104614 |
| Export: | BibTex | RIS | EndNote |
Binding Isotherm Simulations
The plot depicts the binding isotherm simulation of a 1:1 interaction of Serotonin (0.0002920134326179004 M) and Zeolite L3.0 (0 — 0.0005840268652358008 M).
Please sign in: customize the simulation by signing in to the SupraBank.